Measurement of Protein Concentration with NanoDrop Spectrophotometer
The NanoDrop spectrophotometer uses surface tension to hold small liquid samples in place, eliminating the need for cuvettes or capillaries. It operates based on UV/Vis spectroscopy, measuring light absorbance at specific wavelengths.
This tutorial provides step-by-step instructions for measuring protein concentration at 280 nm. To do so accurately, you must also know the extinction coefficient and molecular weight of the measured protein, which can be obtained using the ProtParam tool.
📦 Equipment (available at the NanoDrop station)
- NanoDrop spectrophotometer (with NanoDrop GUI)
- Milli-Q detergent (for cleaning)
- Pipettes (1–20 μL range)
- Wiping tissues
🧪 User-Provided Materials
- NMR buffer (for blanking)
- Protein sample
- Extinction coefficient and molecular weight (from ProtParam)
1. Obtaining the Extinction Coefficient via ProtParam
- Visit ProtParam.
- Paste your protein’s FASTA sequence.
- Click
Compute Parameters.
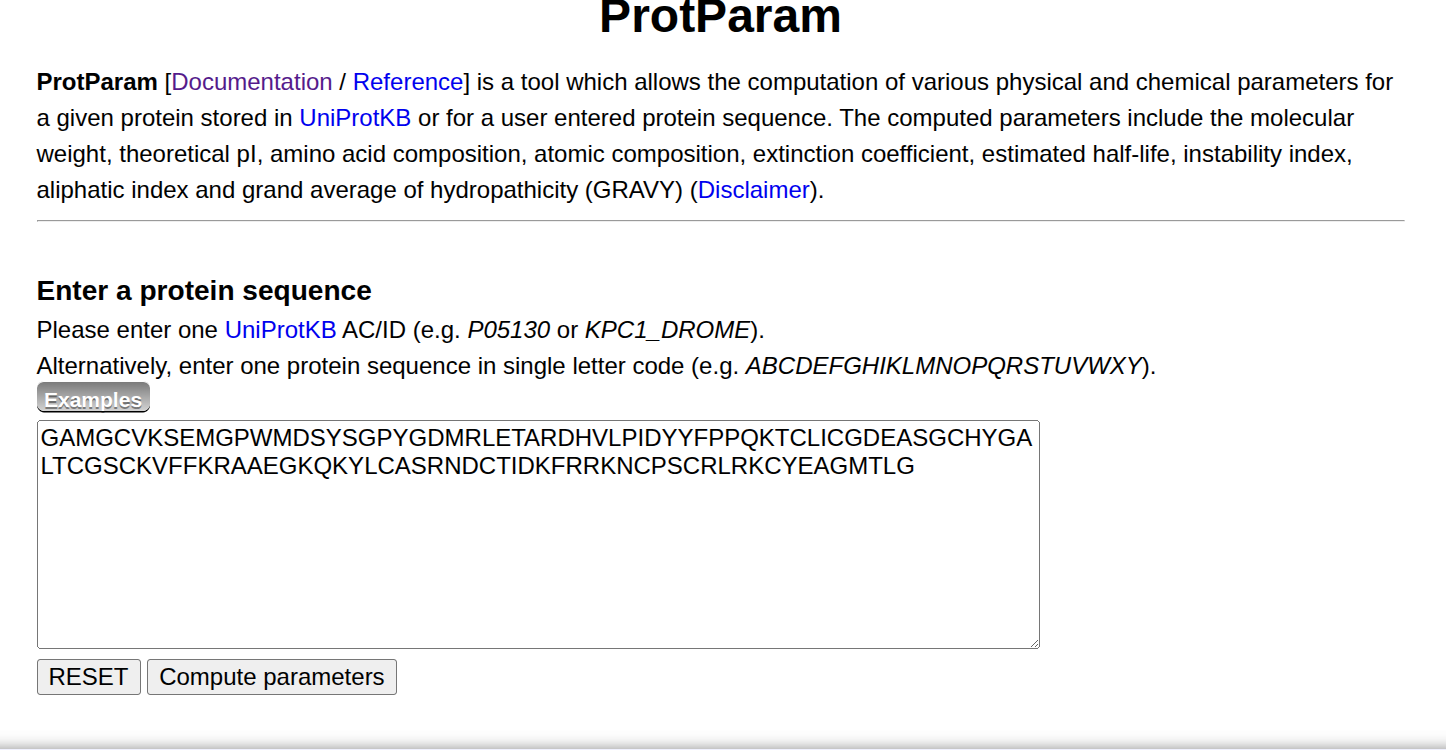
- Locate the Extinction Coefficient section.

- ✅ If your buffer contains reducing agents (e.g., DTT or TCEP), choose the value assuming all Cys are reduced.
2. Preparation of Samples for Measurement
Before performing NanoDrop measurements, the protein solution must be diluted to ensure that the absorbance at 280 nm (A280) falls within the linear range of the Beer–Lambert law, typically between 0.1 and 1.0.
Because the stock concentration is often unknown, begin with a test dilution and increase if necessary. Once you determine the correct dilution factor, prepare a triplicate set for accurate measurement.
📐 Useful Equations
\begin{align*}
A &= c \times \epsilon \times l \\
dilution &= \frac{V_{sample} + V_{buffer}}{V_{sample}} \\
total \ dilution &= dilution_1 \times dilution_2 × ...\\
c [μM] &= \frac{A \times 10⁶ \times dilution}{\epsilon} \\
c [μM] &= \frac{(c_{mg/mL} \times 10⁶ \times dilution)}{MW}
\end{align*}
🔬 Step 1: Prepare a 20× Test Dilution Sample
- Pipette 2 μL protein and 38 μL buffer into a new Eppendorf tube → total = 40 μL
- Dilution = 20×
- Put the vial into vortex and mix well
🧪 Step 2: Measure A280
- Measure 3 μL on NanoDrop (see below).
- If A280 < 1.0, proceed to replicate measurements
- If A280 > 1.0, dilute further according to following table:
| Final Dilution | Dilution Step | Test Dilution Sample | Buffer | Total Vol |
|---|---|---|---|---|
| 30× | 1.5× | 6 μL | 3 μL | 9 μL |
| 40× | 2× | 5 μL | 5 μL | 10 μL |
| 50× | 2.5× | 4 μL | 6 μL | 10 μL |
💡 Example: 20× → 2× → 40× total dilution.
Always mix the new dilution table into a new Eppendorf tube.
✅ Step 3: Prepare Final Replicates
Once you’ve determined the appropriate dilution factor, prepare three independent replicate samples using that same dilution. Each replicate should provide at least 15 μL of solution (sufficient for 5 × 3 μL measurements).
For example, if the final dilution is 30×:
- In three clean Eppendorf tubes, mix 2 μL of protein stock with 58 μL of NMR buffer.
- Vortex each sample thoroughly to ensure proper mixing.
- If the initial 20× test dilution yielded an acceptable absorbance (within 0.1–1.0), you may use it as Replicate 1, and prepare two additional samples following the same 20× protocol.
3. Measurement Procedure
- Clean both measuring tips:
- Open Nanodrop and pipette 3 μL of Milli-Q onto lower tip.
- Close lid briefly.
- Wipe both tips with tissues.
- Open NanoDrop measurement settings:
- On the GUI, Select “Protein A280”.
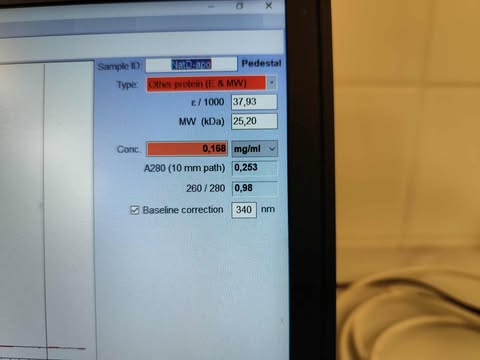
-
Under
Sample ID, name your measurements. -
Under
Type, choose:- “Other protein (E & MW)”
- Enter ε from ProtParam (in thousands, e.g., 21 for 21,000), using the value that assumes all Cys residues are reduced.
- Enter MW in kDa.
- “Other protein (E & MW)”
- Blank:
- Pipette 3 μL of NMR buffer onto lower tip.
- Close lid, click Blank.
- Wait for the blanking process to finish.
- Wipe both tips with tissues.
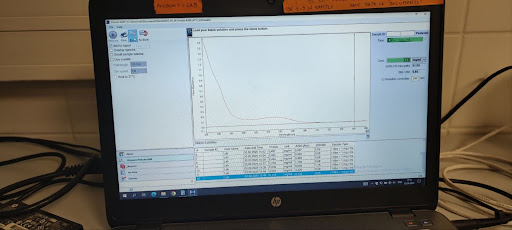
- Measure sample:
- Wipe both tips with tissues.
- Pipette 3 μL of prepared protein sample onto lower tip.
- Close lid, click Measure.
- Wait for the measuring process to finish.
- Wipe both tips with tissues.
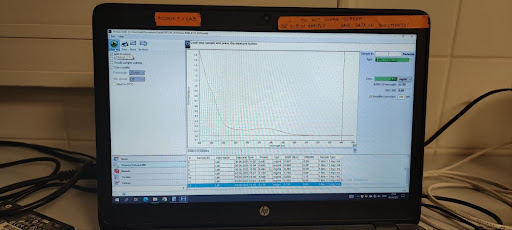
- Record results:
- A280
- Conc. mg/mL value reported by NanoDrop
🧼 Only clean with Mili-Q and return to the homescreen after final replicate, not between every measurement
4. Calculating Protein Concentration
After collecting your NanoDrop measurements, you can calculate the protein concentration in two ways, depending on whether you recorded:
- Absorbance values (A280) → use the Beer–Lambert law
- Mass concentration (mg/mL) reported by NanoDrop → convert to molar concentration
🧠 Recommendation: Record both the absorbance and the reported concentration during measurement. This provides a useful cross-check and allows you to recompute values if needed.
🔹 Option 1: Calculation Based on Absorbance (A280)
If you chose to manually calculate concentration using the absorbance values, follow these steps:
- For each replicate, average the 5 A280 values recorded.
- Apply the Beer–Lambert law:
\begin{align*} A &= c \times \epsilon \times l \\ c~[μM] &= \frac{(A_{avg} × 10^6 × dilution)}{ε} \end{align*}Where:
A= average absorbance at 280 nm (unitless)ε= extinction coefficient in M⁻¹·cm⁻¹ (from ProtParam)l= path length in cm (NanoDrop usually auto-adjusts; use 0.1 cm unless known otherwise)dilution= total dilution factor applied to the measured sample
🔹 Option 2: Calculation Based on NanoDrop Reported Concentration
If you used “Other protein (E & MW)” mode, NanoDrop will report mass concentration in mg/mL.
To convert this to the final molar concentration, use the following formula:
c~[μM] = \frac{c_{NanoDrop} × 10^6 × dilution}{MW}
📝 Note: The two methods should give you very similar results. If they differ significantly, re-check your ε, MW, dilution factor, and pipetting accuracy.
✅ Final Result
Once you’ve calculated the molar concentration for all three replicates:
- Average the three values to obtain your final concentration estimate.
- If necessary, report the standard deviation for accuracy.
🧠 Tips & Best Practices
- Always mix dilutions thoroughly
- Do not rely on “1 Abs = 1 mg/mL” mode
- Clean tips carefully — but only do a full reset after the full triplicate
- Use both A280 and mg/mL values when possible
- Never forget to apply the dilution factor